A-Eye
// AI that helps healthcare professionals diagnose eye disease
A-Eye: Architecture of a Deep Learning Pipeline for Ophthalmological Diagnosis
In the critical field of medical imaging, an algorithm’s accuracy alone is not sufficient; robustness and explainability are essential. A-Eye was born from this requirement.
It is an artificial intelligence solution designed to assist ophthalmologists in the analysis of retinal fundus images, achieving approximately 99% accuracy on test datasets.
This article details the technical architecture of the project, the challenges of medical image processing, and the internal mechanisms of the convolutional neural networks (CNNs) used.
The Technical Challenge: Normalizing Visual Chaos
The initial dataset (12,000 images from 6,000 patients) exhibited strong heterogeneity: variations in lighting conditions, differing resolutions, and capture artifacts. Before even discussing AI, substantial data engineering work was required.
Standardized Preprocessing:
All images underwent uniform resizing and pixel normalization to accelerate model convergence during training.
Data Augmentation:
To prevent overfitting on a limited medical dataset, we implemented dynamic augmentation techniques: random rotations, zooming, and brightness adjustments. These transformations force the model to generalize disease-related features rather than memorizing specific images.
The Architecture: A Multi-Stage CNN Pipeline
Rather than building a single monolithic model, we opted for a multi-stage pipeline architecture, inspired by data engineering best practices to improve efficiency and modularity.
Stage 1: The Gatekeeper (Image Validation)
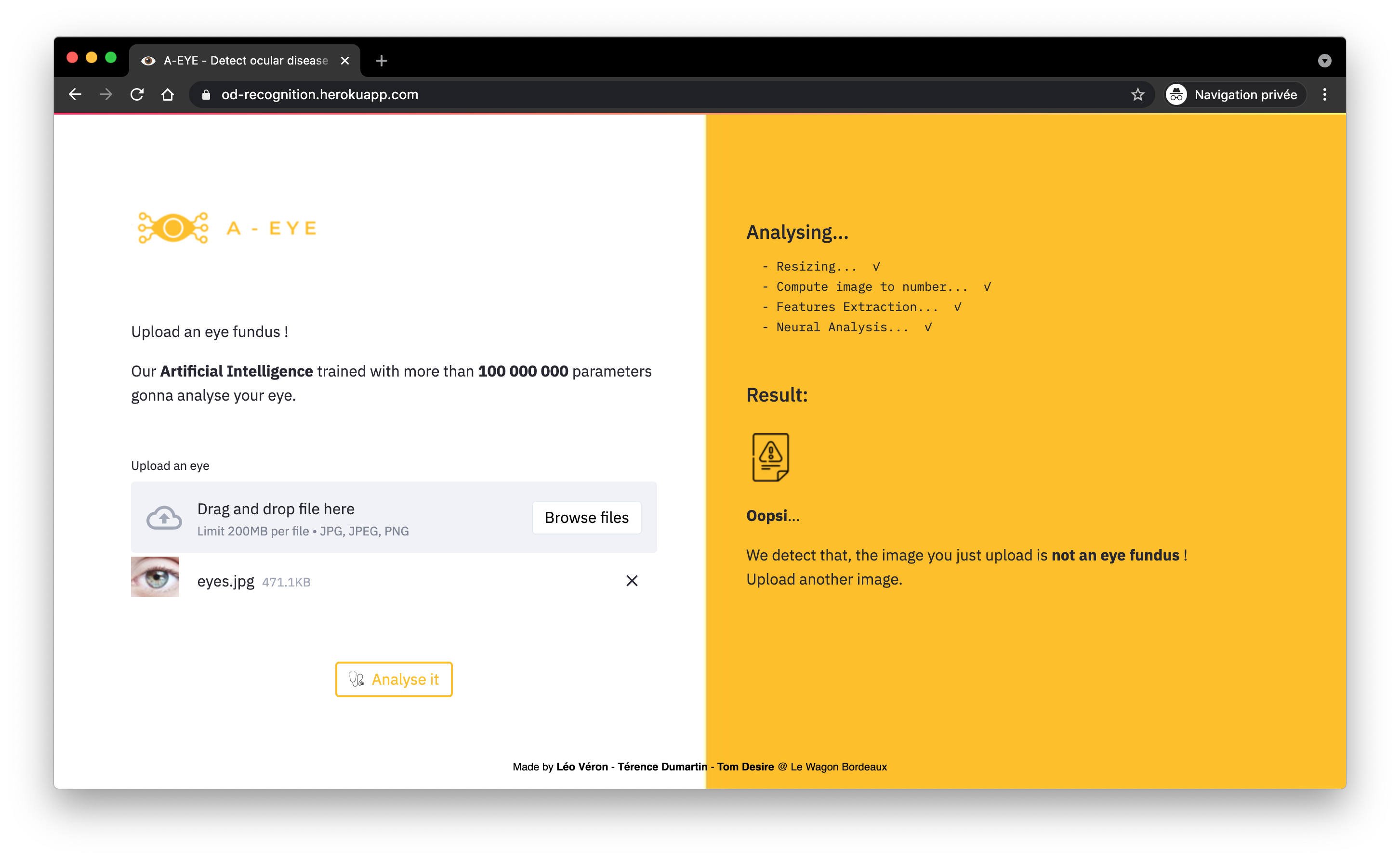
A lightweight CNN acts as a quality filter. Its role is a binary classification: “Usable” vs “Unusable.”
It rejects images that are too blurry or poorly framed, which would otherwise degrade the final diagnosis.
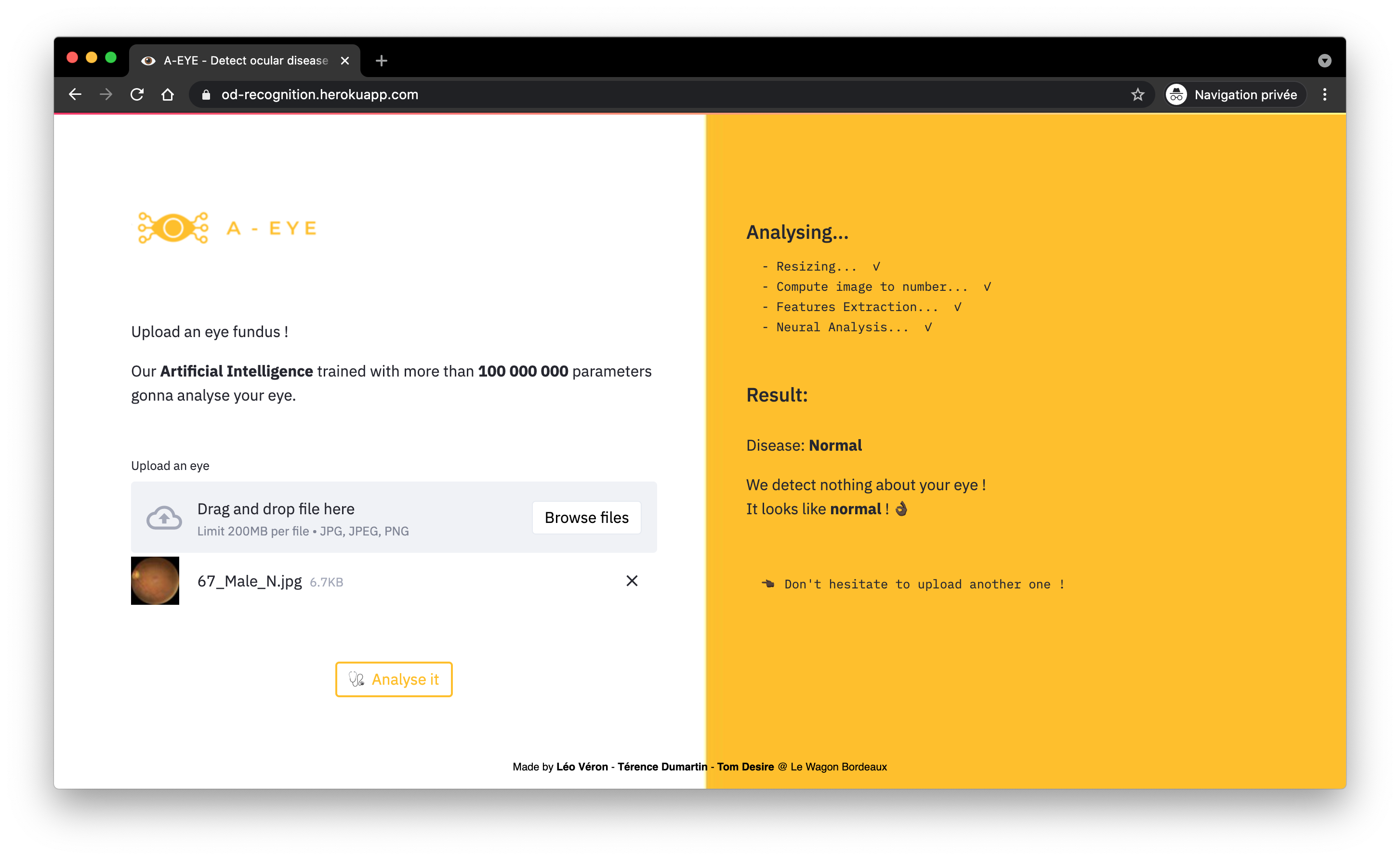
Stage 2: Rapid Triage (Binary Classification)
The core of the system is a model optimized not only for accuracy but primarily for recall (sensitivity).
The objective is to minimize false negatives (missing a diseased eye). This model quickly separates healthy cases from pathological cases requiring immediate attention.
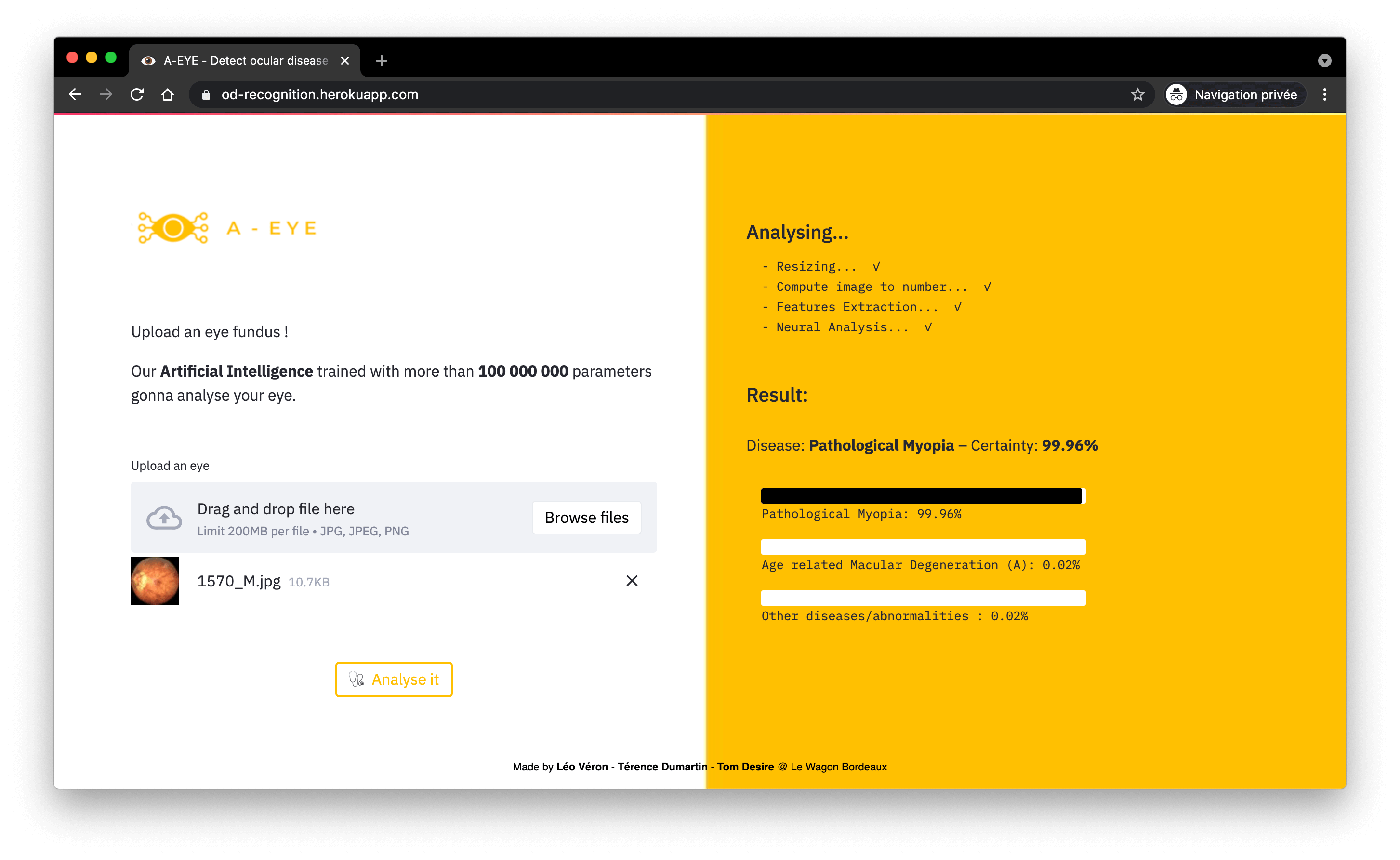
Stage 3: Differential Diagnosis (Multi-Class Classification)
For images identified as pathological, a deeper network is activated to classify the specific disease. This is where computational power is most heavily utilized.
Inside the AI’s Eye: Convolutions and Feature Maps
How does a matrix of numbers (an image from a computer’s perspective) become a diagnosis? This is the power of Convolutional Neural Networks (CNNs).
The Filter Mechanism
Think of a CNN as a series of successive layers. The model slides small filters (convolution kernels) across the entire image.
- The first layers learn to detect very simple concepts: vertical edges, contrast changes, and basic textures.
- Intermediate layers combine these elements to recognize more complex shapes: circles (the macula) and branched lines (blood vessels).
- The final layers assemble these shapes to identify pathological structures: microaneurysms, exudates, or hemorrhages.
Visualizing the Decision: Activation Maps
A crucial aspect of medical AI is interpretability. How can we trust the model?
During inference, the network generates feature maps. Using techniques such as Grad-CAM, we can produce a heatmap that overlays the original image.
If A-Eye diagnoses diabetic retinopathy, the heatmap highlights in bright red the hemorrhagic lesion areas that drove the decision. This allows physicians to instantly validate the AI’s reasoning.
Technological Leverage: Transfer Learning and Fine-Tuning
Training a deep CNN from scratch requires millions of images. With only 12,000 images available, transfer learning is the optimal solution.
We leveraged well-known architectures (VGG16, InceptionV3) pre-trained on ImageNet, a large-scale general-purpose dataset. The approach consists of:
- Freezing the early layers, which already encode universal knowledge of shapes and colors.
- Replacing and fine-tuning only the final fully connected layers using our domain-specific medical data.
From R&D to Production
To ensure that this pipeline did not remain a purely experimental project, industrialization was considered from the outset:
- Containerization (Docker): The entire pipeline—Python dependencies and large models included—is encapsulated in a Docker image, ensuring reproducibility across any cloud server or local machine.
- Interface (Streamlit): A lightweight web application allows practitioners to interact with the model via simple drag-and-drop, fully abstracting the complexity of the deep learning backend.
Video of the final presentation of our project at Le Wagon
Date
2021
Tech Stacks
- Python
- Github
- MlFlow
- Google Cloud Platform
- Streamlit
- Docker
- Heroku
Team
- Léo Véron
- Tom Desire
- Térence Dumartin